The MotifMap system provides comprehensive maps of candidate regulatory elements encoded in the genomes of model species using databases of transcription factor binding motifs, refined genome alignments, and a comparative genomic statistical approach- Bayesian Branch Length Score
Select species track and click next
Select motif and click next
| Motif ID | TF Name | Consensus | Length |
|---|
->
Set filters and click search to list binding sites
| cid | Location | +/- | BBLS | BLS | NLOD | Z-Score | FDR | Closest Gene | Distance (bp) | Description |
|---|
Select species (alignment) and click next
Enter a gene name (official gene symbol or refSeq transcript id) and click next
->
Set filters and click search to list binding sites
| cid | Location | +/- | BBLS | BLS | NLOD | Z-Score | FDR | Motif ID | TF Name | Gene | Distance (bp) | Region |
|---|
Select species (alignment) and click next
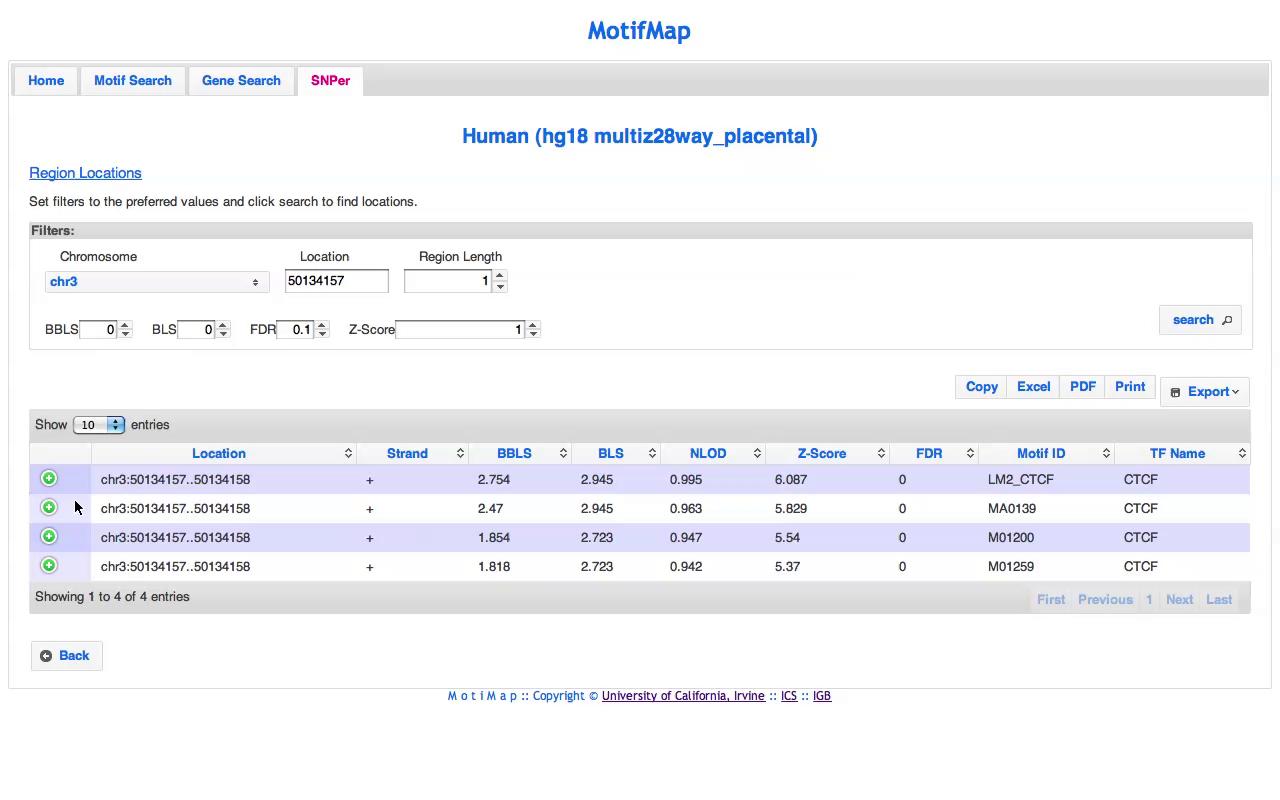
Select genome location and click search
| cid | Location | Strand | BBLS | BLS | NLOD | Z-Score | FDR | Motif ID | TF Name |
|---|